Loom
LOOM format
ASAP accepts as input HDF5 files following the LOOM format, developped by Linnarson lab. Note that ASAP complies with 3.0.0 specifications.
10x
10x H5 format
TXT
Plain-text format (compressed or not)
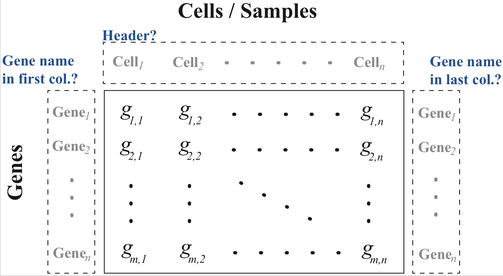
ASAP accepts as input simple Plain-text matrices, with as column delimiter tabulation, comma, semi-colon or space.
First or last column as gene names is optional. Gene names are then mapped to the Ensembl database.
First line as cell/sample names is optional too.
The file can be compressed.

TAR
Archive
ASAP supports archive files that contain several datasets in a plain-text format. You can select which dataset to take as input in the interface.
TGZ
Archive compressed
ASAP supports compressed archive files that contain several datasets in a plain-text format. You can select which dataset to take as input in the interface.
ZIP
Compressed
ASAP supports compressed files (zip or gzip).
MEX
MEX/MTX
ASAP requires all three files to be archived/compressed in a single file (.tar, .tar.gz, .gz, .zip, .bz2, .tar.bz2). The files need to be named:
- barcodes.tsv
- genes.tsv or features.tsv
- the main data matrix in a file with extension .mtx. If several mtx files are present, a file named matrix.mtx is required and only this one will be used
RDS
RDS
From version 8, ASAP supports Seurat objects (rds).