Tutorial 3 : How to import data from 10x [from CellRanger output]
In this tutorial we will show you how to find/retrieve a 10x Genomics dataset from the output of CellRanger and upload it to ASAP.
Find the necessary file (.h5 file)
After running CellRanger on your 10x library, you should obtain a folder containing many different files.
These files are better detailed on 10x main website:
For ASAP, the only important files are:
- Filtered feature-barcode matrices HDF5: /home/jdoe/runs/sample345/outs/filtered_feature_bc_matrix.h5
- Unfiltered feature-barcode matrices HDF5: /home/jdoe/runs/sample345/outs/raw_feature_bc_matrix.h5
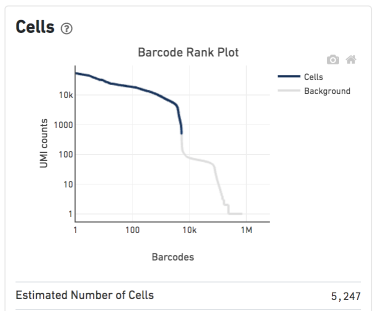
For ASAP, it makes more sense to use the filtered HDF5 file, as all empty droplets were already filtered out. The only exception would be in the case you don't like the filtering that was perfomed by CellRanger, and you prefer to do it in ASAP (see Figure 1). For example if the curve seems to threshold before an interesting subpopulation, or if the number of cells detected is very different from what you estimated.
In this case, you should know that the parsing/filtering steps in ASAP will take much more time, because the number of cells will probably be close to 1M. So it's only recommended to do this in case the output of CellRanger looks weird/incorrect to you.
Of note
CellRanger also output other stats/plots such as t-SNE and clusterings. However these results cannot be imported in ASAP, because they are not stored within the .h5 file.
Upload the .h5 file to ASAP
ASAP can take .h5 files from 10x directly as input file.
Simply create a "New Project" from the main page, and upload your .h5 file.
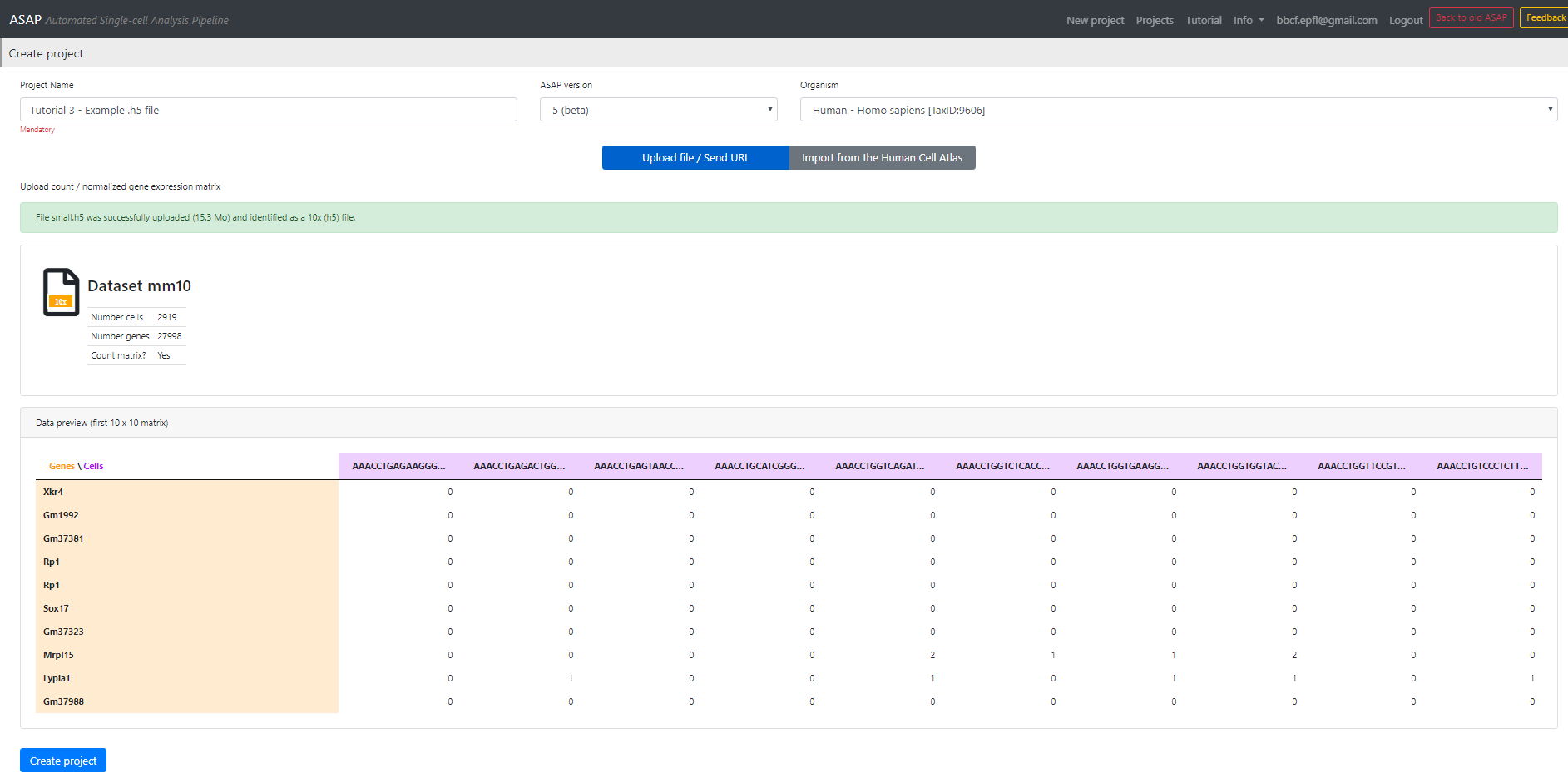
Once uploaded, you will see a preview of the file (see Figure 2) with first 10 cols/ 10 rows, row names, cell barcodes, number of cells, number of genes, ...
If it looks correct to you then you can click on [Create Project].
Run a pipeline in ASAP
Once this is done, you can process your dataset the same way than with any other input file.
Don't hesitate to visit the Tutorial 2 for a detailed explanation of how to run a project in ASAP.